Structural basis of defects in the sacsin HEPN domain responsible for autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS).
Kozlov, G., Denisov, A.Y., Girard, M., Dicaire, M.J., Hamlin, J., McPherson, P.S., Brais, B., Gehring, K.(2011) J Biol Chem 286: 20407-20412
- PubMed: 21507954
- DOI: https://doi.org/10.1074/jbc.M111.232884
- Primary Citation of Related Structures:
3O10 - PubMed Abstract:
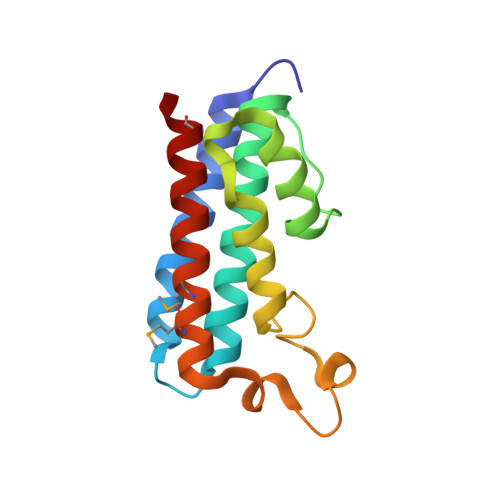
Sacsin is a 520-kDa protein mutated in the early-onset neurodevelopmental and neurodegenerative disease autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS). The C terminus of the protein contains an HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domain of unknown function. Here, we determined the high-resolution 1.9-Å crystal structure of the HEPN domain from human sacsin. The structure is composed of five parallel α-helices with a large loop of several short helical segments. Two HEPN protomers assemble as a dimer to form a large positively charged cavity at the dimer interface that binds GTP and other nucleotides. The crystal structure reveals that the ARSACS N4549D mutation disrupts dimerization and protein folding. This study provides novel insights into the oligomerization state of sacsin and functions that are lost in mutations that cause ARSACS.
Organizational Affiliation:
Department of Biochemistry, Groupe de Recherche Axé sur la Structure des Protéines, McGill University, Montréal, Québec H3G 0B1, Canada.